David Reid Jacobson, Ph.D.Current:Postdoc, JILA, NIST and CU BoulderStarting January 2023:Assistant Professor, Department of Chemistry, Clemson UniversityCurriculum Vitae |
 |
| In January 2023, I will be starting my own lab in the Department of Chemistry at Clemson University, using single-molecule force-application techniques to study the physical chemistry of membrane proteins. Please contact me if you are interested in joining the group. Currently, I am a postdoctoral researcher in the lab of Tom Perkins at the University of Colorado, using atomic force microscopy to study the model membrane protein bacteriorhodopsin. This, and my prior work, is described below. |
 |
Follow me on Mastodon: @Jacobson@fediscience.org
Membrane protein folding (CU Boulder)
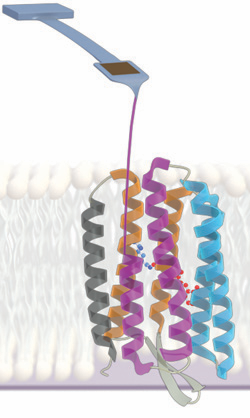 |
I am currently using atomic force microscopy (AFM) to measure the energetics stabilizing membrane proteins, especially the model membrane protein bacteriorhodopsin. In my first published contribution to this area, I developed a new technique (dubbed "zigzag force spectroscopy") to extract more usable information from each individual molecule probed compared to traditional techniques. (Featured as a "New and Notable" article in Biophysical Journal) With Hao Yu, I then used AFM-based unfolding to measure the unfolding free energy of an eight-amino-acid region of bacteriorhodopsin, overcoming the limitations present when such a measurement is made using the traditional approach of chemical denaturation in detergent micelles. In the course of this work, I developed an analytical theory for correcting unfolding/refolding kinetic rates in cases where the AFM time resolution is limiting. I then extended this approach to measure the change in unfolding free energy when a single-amino-acid point mutation is introduced. This reports on the contribution of the mutated amino acid's side chain to the overall stability of the protein. Using single-molecule techniques opens an avenue for making such measurements even when traditional biochemical assays cannot be applied. Left: Illustration of an experiment in which mechanical force is used to unfold bacteriorhodopsin. The force needed to disrupt the native structure reports on the underlying stability of that structure. |
Nucleic acid electrostatics (UCSB)
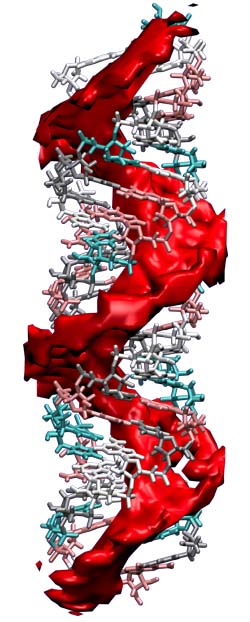 |
During my Ph.D. (2011-2016) in the lab of Omar Saleh, I used magnetic tweezers force spectroscopy, bulk dialysis, and theoretical methods to study how the strong negative charge of DNA and RNA affects their solution conformations and interactions with ions in salt solution. Review article: "Counting the ions surrounding nucleic acids" Main results: "Single-stranded nucleic acid elasticity arises from internal electrostatic tension" "The snakelike chain character of unstructured RNA" Technical/theoretical papers: "Magnetic tweezers force calibration for molecules that exhibit conformation switching" "A thin permeable-membrane device for single-molecule manipulation" Left: Illustration of the RNA ion atmosphere. The red is the region, based on Poisson-Boltzmann calculation, where the local cation concentration is more than ten times the bulk value of 100 mM. |
Measurement of radon binding to a molecular host (Penn)
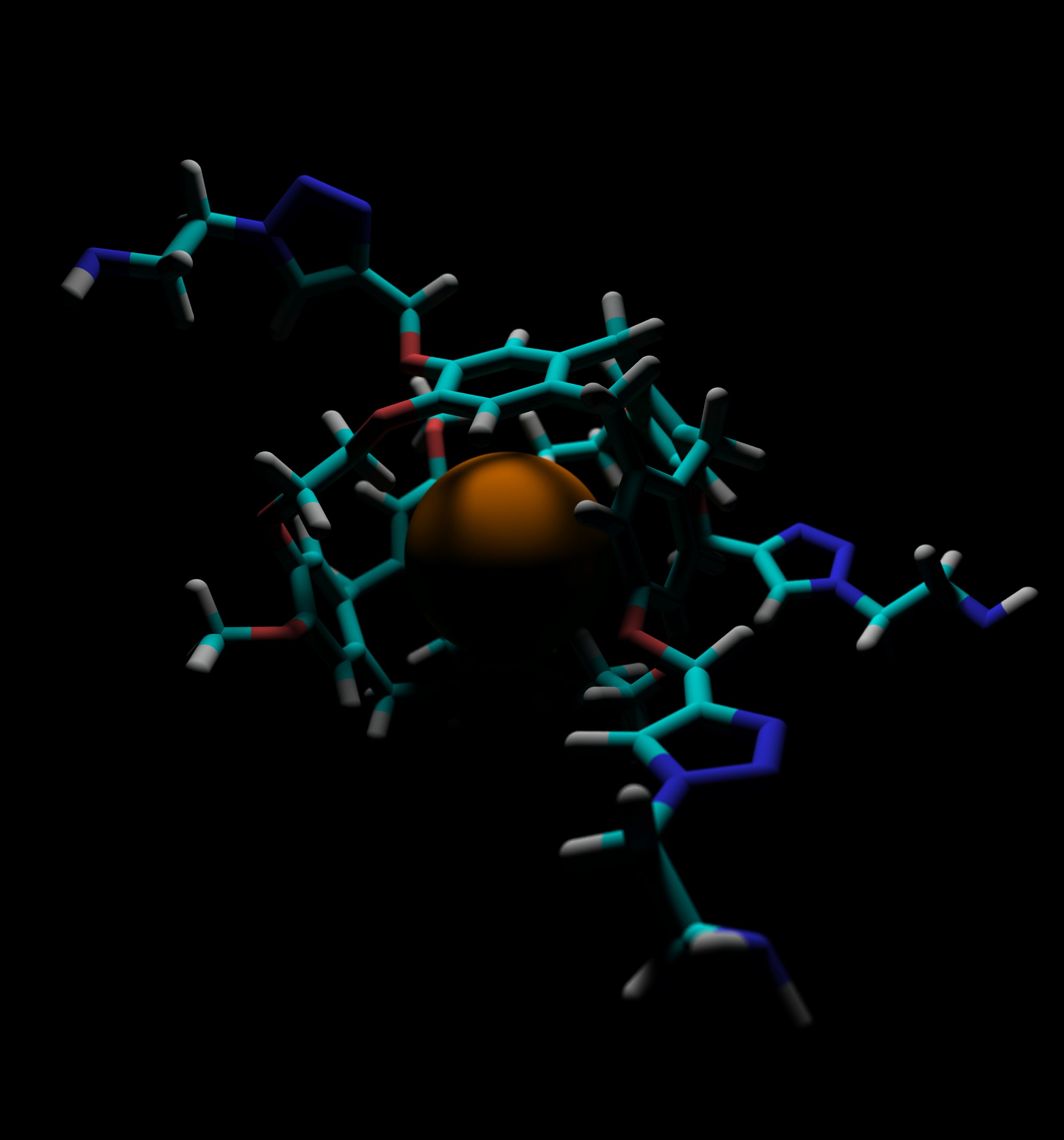 |
From 2009-2011, I worked in the lab of Ivan Dmochowski on a series of projects in the field of supramolecular chemistry. Most sucessfully, in collaboration with Ron Colle at NIST (Gaithersburg, MD), I developed a method for measuring the binding affinity of radon to a discreet molecular species. Resulting publications: Left: Depiction of the inert radon atom encapsulated within the TTEC cyrptophane molecule. |
The Atacama Cosmology Telescope (Penn)
 |
During the summer of 2008, I worked in the lab of Mark Devlin on ACT, including having the opportunity to travel to the telescope site in northern Chile. Left: ACT, at an elevation of more than 17,000 ft above sea level, as seen from the slopes of Cerro Toco. |